I’m a fifth year PhD candidate in the Bo Wang Lab, Department of Medical Biophysics, at the University of Toronto and Vector Institute. I’m also co-supervised by Kieran Campbell and Michael D. Taylor.
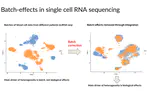
My research involves the development and application of machine learning and computational biology methods for genome sequencing data. During my PhD, I have worked on representation learning for single-cell sequencing, spatial transcriptomics, and histopathology data, for tasks such as integration, predictive modeling, and dynamic modeling. I have also worked with various groups performing single-cell analysis in development and cancer biology settings. More broadly, I am interested in advancing our understanding of human disease through the lens of genome sequencing, computational biology, and machine learning.
I’ve previously interned at Microsoft Research (x2), where I worked on the Antigen Map Project, and The Vector Insitute’s AI Engineering Team where I extended a COVID-19 genome analysis tool that I developed.
Download my CV to see full details of my academic and industry experience.
See my Google Scholar for a complete list of publications.
I have a photography website at shotsby5.com where I share photographs of landscapes, cities, and hopefully astrophotography soon.
Please get in touch if you’d like to chat about research and/or collaboration!
- Computational biology
- Single-cell genomics
- Spatial transcriptomics
- Histopathology
- Representation learning
- Large language models
PhD in Medical Biophysics, 2020-
University of Toronto
Master of Bioinformatics (M.Binf), 2019
University of Guelph
Bachelor of Science (BSc.), 2017
University of Waterloo