A Deep Learning Framework for Estimating Cell-specific Kinetic Rates of RNA Velocity
Abstract
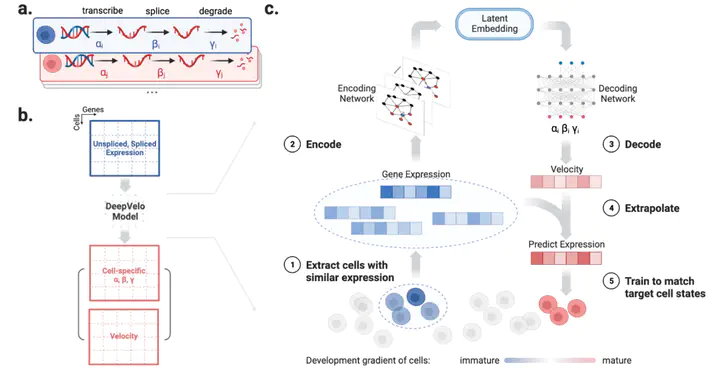
Existing RNA velocity estimation methods rely on strong assumptions of predefined dynamics and cell-agnostic constant transcriptional rates, which are often violated in complex and heterogeneous single-cell RNA sequencing (scRNA-seq) data. To overcome these limitations, we present DeepVelo, a novel method that estimates cell-specific dynamics of transcriptional kinetics using a Graph Convolution Neural Network (GCN) model and generalizes RNA velocity to cell populations containing time-dependent kinetics and multiple lineages. We applied DeepVelo to complex developmental datasets, including dentate gyrus and hindbrain neurogenesis, and demonstrated its ability to disentangle multifaceted kinetics. DeepVelo infers time-varying cellular rates of transcription, splicing and degradation, recovers each cell’s stage in the underlying differentiation process, and detects putative driver genes regulating these processes. By relaxing the constraints of previous techniques, DeepVelo facilitates the study of more complex differentiation and lineage decision events in heterogeneous scRNA-seq data. The DeepVelo package is available at https://github.com/bowang-lab/DeepVelo.