Disrupted chromatin insulated interactions of long non-coding RNAs and protein-coding genes indicate transcriptional rewiring of oncogenic and tumor-suppressive pathways
Abstract
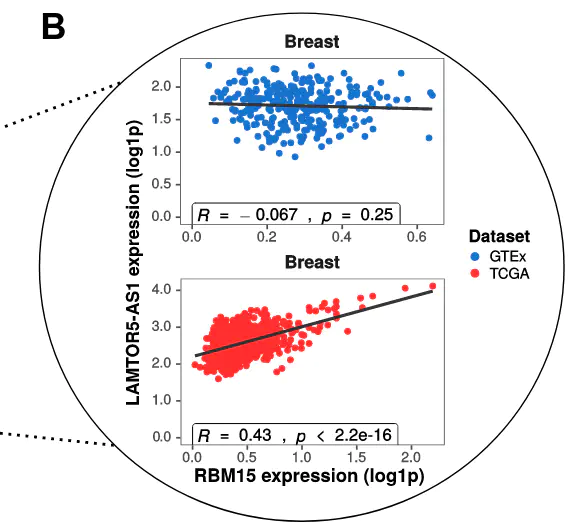
Long non-coding RNAs (lncRNAs) have diverse roles in gene regulation, but remain poorly annotated in the context of healthy human tissues and cancer. Transcriptional gene regulation involves interactions of transcription factors with gene promoters and distal enhancers, mediated by three-dimensional genome architecture. Enhancers are often transcribed, and enhancer RNAs have significant classification overlap with lncRNAs. Here, we analyzed 7277 cis-regulatory modules frequently bound by diverse transcription factors that encoded 1057 lncRNAs and interacted with protein- coding gene promoters through chromatin loop insulation across at least 5 unique Hi-C datasets. To analyze expression patterns of these interactions, we determined 11 pairs of tumor tissues and corresponding healthy tissues by annotating transcriptomics datasets from The Genotype-Tissue Expression Project (GTEx) and The Cancer Genome Atlas (TCGA). Stringent analysis of variance (ANOVA) tests were performed to identify significant putative regulatory links between lncRNAs and promoter regions in a tissue-specific manner. We identified 444 unique tissue-specific significant interactions between lncRNAs and promoters of protein-coding genes. Most significant interactions were present in tumor tissue (413) and much fewer in healthy tissue (113), indicating the formation of oncogenic regulatory interactions in cancer cells through remodelling of chromatin architecture. Conversely, 31 significant links in healthy tissue were non-significant in tumor tissue, suggesting the breakdown of tumor-suppressive interactions. Protein-coding genes in TCGA or GTEx exclusive significant interactions were enriched in cancer-related pathways such as DNA repair, apoptosis regulation, angiogenesis, and cell cycle regulation. We identified key disruptions in esophageal carcinoma involving tumor-suppressors RNF43 and FBXO11, and their respective associations with lncRNAs BZRAP1-AS1 and AC079807.2. Our results corroborate previous discoveries of lncRNA mechanisms of gene regulation mediated by chromatin loops and three-dimensional genome architecture. We demonstrate a broad network rewiring of these interactions in tumor-tissue through changes in chromatin structure and identify putative oncogenic links that may have prognostic and therapeutic potential.